 |
| Asst. Prof. Dr. ERSIN EMRE OREN
Department of Biomedical Engineering
Department of Materials Science and Nanotechnology Engineering
TOBB University of Economics and Technology
Sogutozu Cad. No: 43,
Sogutozu, Ankara, 06560 TURKEY
e-mail: eeoren@etu.edu.tr
phone: +90 (312) 292-4514 (office); +90 (312) 292-4585 (lab)
fax: +90 (312) 292-4091 |
 |
|
|
|
 |
RESEARCH HIGHLIGHTS
Computational science and engineering is a multidisciplinary field that uses sophisticated algorithms and high-performance computers to understand and solve complex problems especially in the field of bionanotechnology. The work in this area enables us both to reduce the budget and time spent on experimental work and to gain new knowledge by simulating some experiments that cannot be done with today's technologies. Nowadays, many inventions made in the fields of bio and nanotechnology are possible due to theory/simulations. In TOBB ETU Bionanodesign (BNT) Laboratory, the problems encountered in bionanotechnology are defined by differential equations / atomic approaches and then algorithms are developed for numerical solutions. By using these techniques developed, various problems encountered in biotechnology and nanotechnology fields (thin film growth kinetics, electromigration, sintering, organic / inorganic interactions, protein / DNA structures, ballistic heat conduction etc.) are investigated and new molecular systems (drug, peptide, DNA) are designed. |
 |
|
Design of Quantum Dots
Quantum dots (QDs), metallic or semiconductor nanoislands, have discrete energy levels thus a well-defined band gap, which may be engineered by controlling their size and morphology. Among several production techniques, Stranski Krastanov (SK) growth mode leads to the formation of dislocation-free nanoislands, which are interconnected with a thin wetting layer, which is used to harvest the photo-excited charge carriers. These unique features make QDs promising candidates for designing novel electronic, magnetic, photonic and biotechnological, devices with improved performance and reliability. In TOBB ETU Bionanodesign (BNT) Laboratory, we modeled the formation, spontaneous evolution and stability of quantum dots during heteroepitaxial growth using various material and process parameters (crystallographic orientation and initial thickness of the film, diffusion and surface stiffness anisotropies, surface and interfacial energies, wetting contact angle, mismatch stresses and external electric and stress fields). The generated information will provide design capability for QDs and hence desired QD-based device technologies. |
|
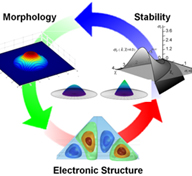 |
|
Antiviral Drug Resistance & Drug Design
Viruses are infectious agents, which can only reproduce in living cells, and can cause epidemics and pandemics ending up with considerable number of morbidity and mortality each year. Seasonal influenza A (H1N1) virus is one of the most frequently circulating strains among humans. This virus is resistant to adamantanes group of antiviral drugs (e.g. amantadine and rimantadine), yet it is still susceptible to oseltamivir and zanamivir. These drugs block the activity of the Neuraminidase (NA) enzyme and prevent new viral particles from being released. However, due to continuing evolution, these viruses have been changing in such a way that the antiviral drugs are less effective in treating or preventing illnesses. In TOBB ETU Bionanodesign (BNT) Laboratory, we first analyze the reported (fludb, Influenza Research Database) NA amino acid sequences and generate mutation maps showing those amino acids more susceptible to mutations. Then using this maps, we predict the mutant strains that may start circulating among us in the future. Later, we predict the molecular structure of NA enzyme of these mutant viruses to analyze the efficacy of our current drug arsenal. By knowing the molecular structures of both the mutant NA and the drug, we used molecular docking algorithms to predict the binding site and the drug efficacy. Based on these calculations, we revealed several mutations, which may cause the virus gain resistance to oseltamivir and/or zanamivir. This information will provide us to analyze the severity of future epidemics quickly and may allow us to design novel drugs in advance. |
|
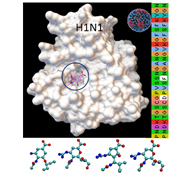 |
|
DNA/RNA/Peptide Based Molecular Electronics
Molecular electronics (ME) is the design and manufacture of electronic devices using molecular building blocks such as DNA/RNA/peptides. This research area is expected to provide the necessary reseurces to extend Moore's Law beyond the foreseen limits of small-scale conventional silicon integrated circuits. To be able to this, the electronic band structure and electronic transport properties of various molecular systems must be understood. DNA is a promising molecule for molecular electronics based on its unique electronic and self-assembly properties. In order to control DNA conductance, its electronic properties have been studied over the years both theoretically and experimentally. Environmental, conformational, and chemical factors exert a large influence on the conductance, making it challenging to control. In TOBB ETU Bionanodesign (BNT) Laboratory, the molecular structures of DNA/RNA in different solutions, having various pH levels and counter ions, are investigated by molecular dynamics techniques. Later the wave functions near the HOMO-LUMO gap are inspected using quantum mechanical techniques such as Density Functional Theory (DFT). |
|
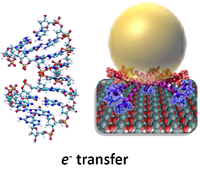 |
|
Design of Drug Release Systems
Controlled-release drug delivery has many advantages compared to conventional single dose drug therapies. Releasing drug molecules in a controlled manner allows us to maintain the drug concentrations within therapeutic range over extended periods, to diminish side effects caused by concentration extremes and repeated administrations, and to improve patient compliance. In spite of these potentials, the use of controlled drug release systems (CDRSs) is limited due to lack of comprehensive understanding on the release kinetics. The release kinetics depends on hydrolysis, diffusive transport of water and drug, polymer erosion, evolution of pore structure, and chemistry, physical size and geometry of the polymer matrix. In TOBB ETU Bionanodesign (BNT) Laboratory, we model the drug release systems by regarding three main mechanisms: diffusion of water through polymer matrix, hydrolysis of drug from polymer matrix and the drug metabolism of human body. To do this, we developed the necessary differential equations with proper boundary conditions and solved these equations by using various numerical techniques and we carried out simulations to understand the relative importance of various kinetic parameters. These results allow us to generate ternary phase diagrams among the three main mechanisms. These ternary phase diagrams provide us to design the geometry and the initial drug load for a given set of drug-polymer matrix systems to sustain the desired therapeutic periods with reduced side effects. |
|
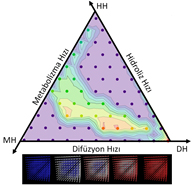 |
|
Protein - Inorganic Interactions
In nature, proteins control nucleation, growth, morphology, crystallography, and spatial organization of minerals and provide molecular scaffolds in the formation of hard tissues with complex and highly functional architectures. Taking advantage of the knowledge, nature has been optimizing over millions of years, we can now understand, engineer, and control peptide-material interactions and exploit them as a new design tool for novel materials and systems with unprecedented functionalities, fabricated using environmentally-benign techniques. Thus, there is a significant promise in harnessing the interface between the inorganic matter (such as minerals, metals, semiconductors) and biologically-derived soft-matter (such as peptides, proteins and DNA). In TOBB ETU Bionanodesign (BNT) Laboratory our project involves data mining (within the publicly available sources such as pdb and pepbank2), developing computer algorithms for the knowledge-based design of functional peptides, and their computational structural analysis in solution and on the inorganic surfaces. |
|
 |
|
Knowledge Based Design
The molecular biomimetics research area is in the crossroads of engineering and life sciences. Gaining insight from evolutionary biology. In nature, biological molecules that perform similar functions usually have similar structures and sequences due to evolutionary, biochemical, and biophysical constraints. Founded on this observation, In TOBB ETU Bionanodesign (BNT) Laboratory, we develop novel bioinformatics approaches to classify peptides based on their functions. This approach performs all-against-all comparisons of peptide sequences categorized for their function and scores the alignments using sequence similarity scoring matrices. We also generate novel specialized similarity matrices that optimize the similarities within the functional peptide sequences and the differences between the functional and non-functional peptide sequences. Using the similarity matrices thus generated, it has been demonstrated that it is possible to computationally design novel peptides with specific functionalities. |
|
 |
|
Electromigration in Integrated Circuits
Electromigration is generally considered to be the result of momentum transfer from the electrons, transported under the applied electric field, to the ions which make up the lattice of the interconnect material. Modern semiconducting chips include a dense array of narrow, thin-film metallic conductors that serve to transport current between the various devises on the chip. These metallic conductors are called interconnects. As integrated circuits become progressively more complex, the individual components must become increasingly more reliable if the reliability of the whole is to be acceptable. However, due to continuing miniaturization of ultra large scale integrated (ULSI) circuits, thin-film metallic conductors or interconnects are subject to increasingly high current densities. Under these conditions, electromigration can lead to the electrical failure of interconnects in relatively short times, reducing the circuit lifetime to an unacceptable level. It is therefore of great technological importance to understand and control electromigration failure in thin film interconnects. In TOBB ETU Bionanodesign (BNT) Laboratory, we developed mathematical models on the mass flow and accumulation on void surfaces, under the action of applied electrostatic and elastostatic force fields, and capillary effects. This formalism follows an irreversible but discrete thermodynamic formulation of interphases and surfaces by taking into account in a natural way the mass transfer process the void growth, between bulk phase and the void region in multi-component systems, in terms of the normalized local values of Gibbs free energy of transformation with respect to the specific surface Gibbs free energy. |
|
 |
|
|
|
|
|
 |
 |
 |
 |
 |
|